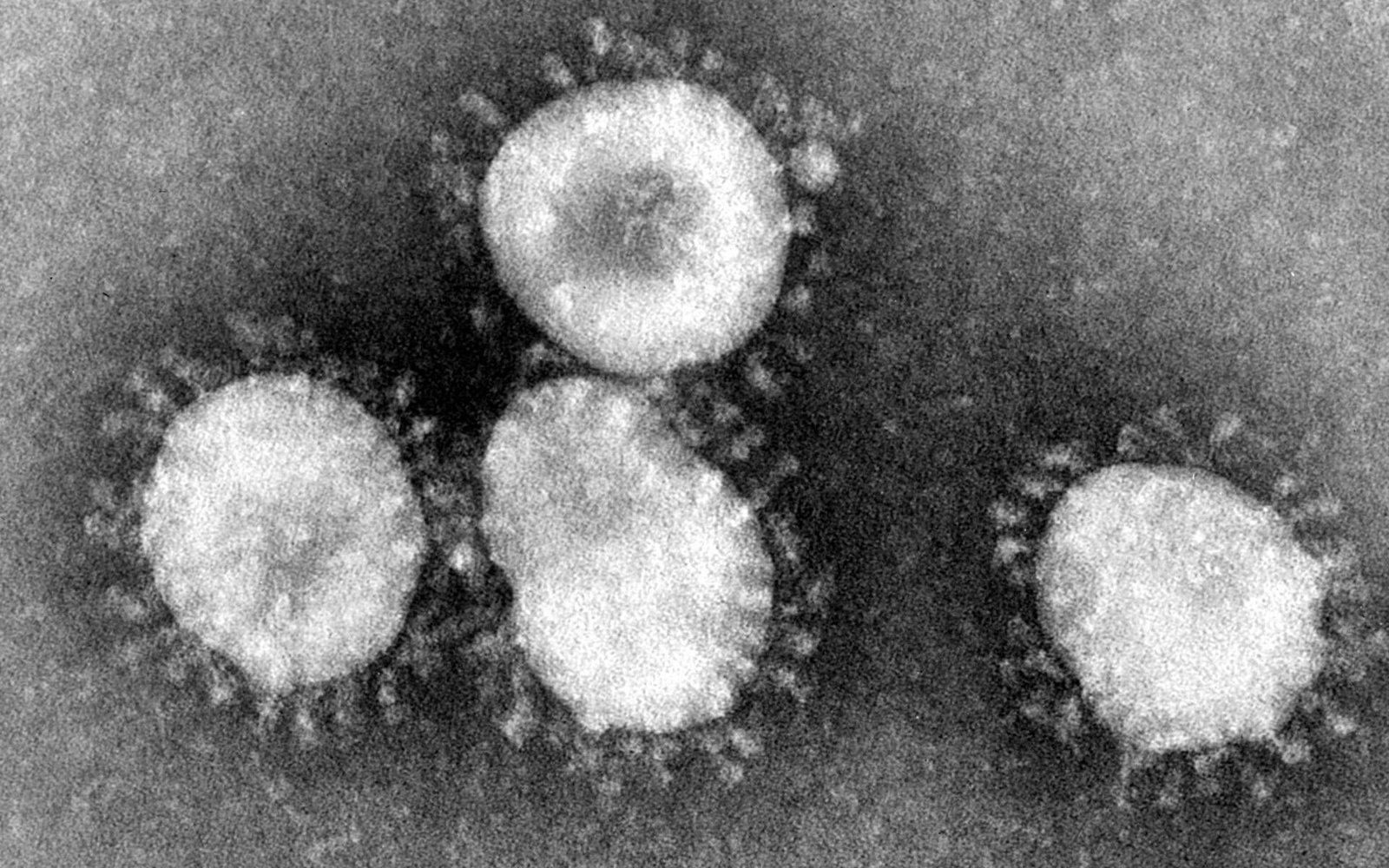
Human coronaviruses (HCoV) are important causes of upper respiratory infections in humans of all ages. In addition, HCoVs have occasionally been associated with pneumonia, meningitis and diarrhoea. HCoV RNA has been detected by RT-PCR in up to 40% of adult human brains (Stewart et al., 1992). All together there are 6 strains of Corona Viruses affected in Human beings till date and now its the seventh strain of 2019 Novel Coronavirus (2019-nCoV) had been found from Wuhan ,China

The 6 strains of Corona Virus known are:
- Human Corona Virus 229E (HCoV-229E)
- Human Corona Virus OC43(HCoV-OC43)
- Human Corona Virus NL63 (HCoV-NL63)
- Human Corona Virus (HCoV-HKU1)
- Severe Acute Respiratory Syndrome (SARS-CoV)
- Middle East Respiratory Syndrome Corona Virus(MERS-CoV)
- 2019 Novel Human Corona Virus (2019 nCoV)
Before 2003, there were only 10 coronaviruses with complete genomes available, including only two human coronaviruses, human coronavirus 229E (HCoV-229E) and human coronavirus OC43 (HCoV-OC43). These two human coronaviruses were discovered in the 1960s, with HCoV-229E being a group 1 coronavirus and HCoV-OC43 a group 2 coronavirus [17, 34]. After the SARS epidemic, up to December 2008, 16 novel coronaviruses were discovered and their complete genomes sequenced. Among these 16 previously unrecognised coronaviruses were two more human coronaviruses, human coronavirus NL63 (HCoV-NL63) and human coronavirus HKU1 (HCoV-HKU1) [37,39], ten other mammalian coronaviruses and four avian corona viruses.

HCoV-229E and HCoV-OC43
HCoV-229E
Phylum: Incertae sedis Order: Nidovirales Family: Coronaviridae Genus: Alphacoronavirus Subgenus: Duvinacovirus Species: Human coronavirus 229E
HC0V-OC43
Phylum: Incertae sedis* Order: Nidovirales Family: Coronaviridae Genus: Betacoronavirus Species: Betacoronavirus 1 Subspecies: Human coronavirus OC43
There are two serotypes of HCoV represented by HCoV-229E and HC0V-OC43. HCoV-229E is a member of Coronavirus serogroup I, which also includes porcine transmissible gastroenteritis virus (TGEV) and feline and canine coronaviruses. The viral attachment protein S of HCoV-229E is unlike S proteins of many coronaviruses in serogroup II, in that it is not cleaved during virus assembly, nor does it cause syncytia formation. HCoV (HCoV-229E and HCoV-OC43) have been known for 30 years.
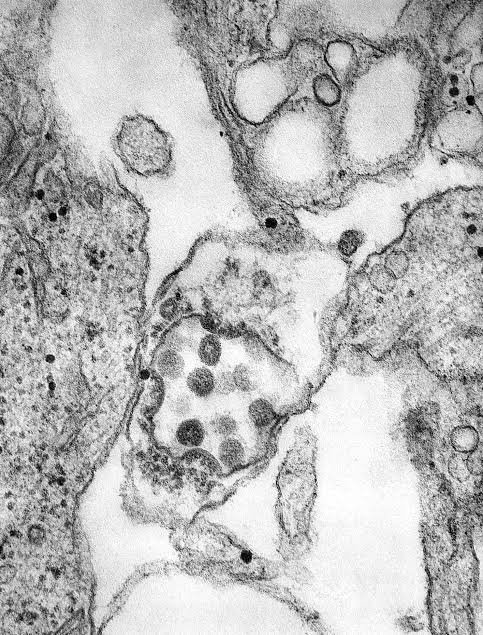
HCoV-229E has been recently involved in nosocomial respiratory viral infections in high-risk children. However, their diagnosis is not routinely performed. Currently, reliable immunofluorescence and cell culture methodologies are not available. As part of a four-year epidemiological study in a Paediatric and Neonatal Intensive care unit, experts have performed and demonstrated the reliability of a reverse transcription-PCR-hybridization assay to detect HCoV of the 229E antigenic group in 2028 clinical respiratory specimens. In hospitalised children (children and newborns) and staff members they found a high incidence of HcoV-229E infection. This reverse transcription-PCR-hybridization assay gave a high specificity and a sensitivity of 0.5 50% Tissue Culture Infective Dose per ml. This technique is reliable and its application for screening large number of clinical samples would improve the diagnosis of HCoVs respiratory infection and our knowledge of these viruses epidemiology.

HCoV-HKU1
Phylum: Incertae sedis Order: Nidovirales Family: Coronaviridae Genus: Betacoronavirus Subgenus: Embecovirus Species: Human coronavirus HKU1
The incidence of HCoV-HKU1 infections is the highest in winter. Similar to other human coronaviruses, HCoV-HKU1 infections have been reported globally, with a median (range) incidence of 0.9 (0 – 4.4) %. HCoV-HKU1 is associated with both upper and lower respiratory tract infections that are mostly self-limiting. The most common method for diagnosing HCoV-HKU1 infection is RT-PCR or real-time RT-PCR using RNA extracted from respiratory tract samples such as nasopharyngeal aspirates (NPA).
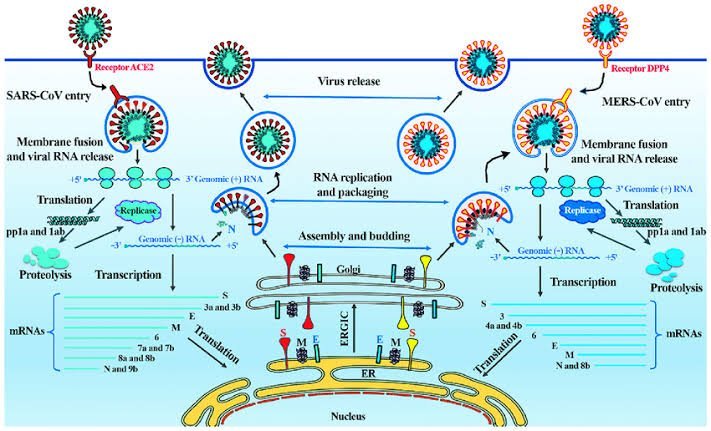
SARS
Severe acute respiratory syndrome (SARS) is a viral respiratory illness caused by a coronavirus called SARS-associated coronavirus (SARS-CoV). SARS was first reported in China (Hong Kong) in February 2003. The illness spread to more than two dozen countries in North America, South America, Europe, and Asia before the SARS global outbreak of 2003 was contained.
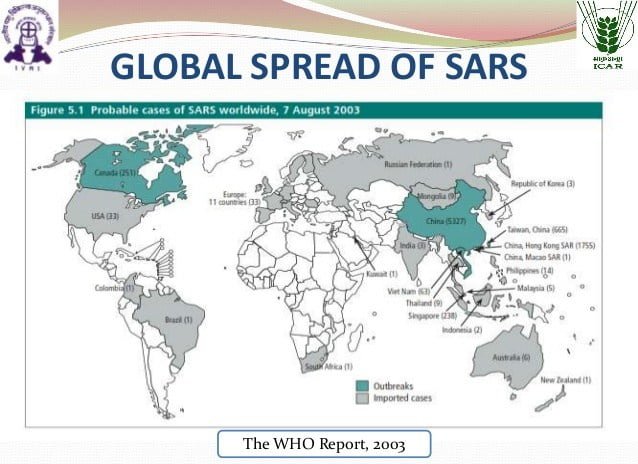
The most typical diseases include, among other things, pulmonary fibrosis, osteoporosis, and femoral necrosis, which have led to the complete loss of working ability or even self-care ability of these cases. Since 2004, there have not been any known cases of SARS reported anywhere in the world. The content in this website was developed for the 2003 SARS epidemic. But some guidelines are still being used.
MERS
Middle East Respiratory Syndrome (MERS) is an illness caused by a virus (more specifically, a coronavirus) called Middle East Respiratory Syndrome Coronavirus (MERS-CoV). Most MERS patients developed severe respiratory illness with symptoms of fever, cough and shortness of breath. About 3 or 4 out of every 10 patients reported with MERS have died.
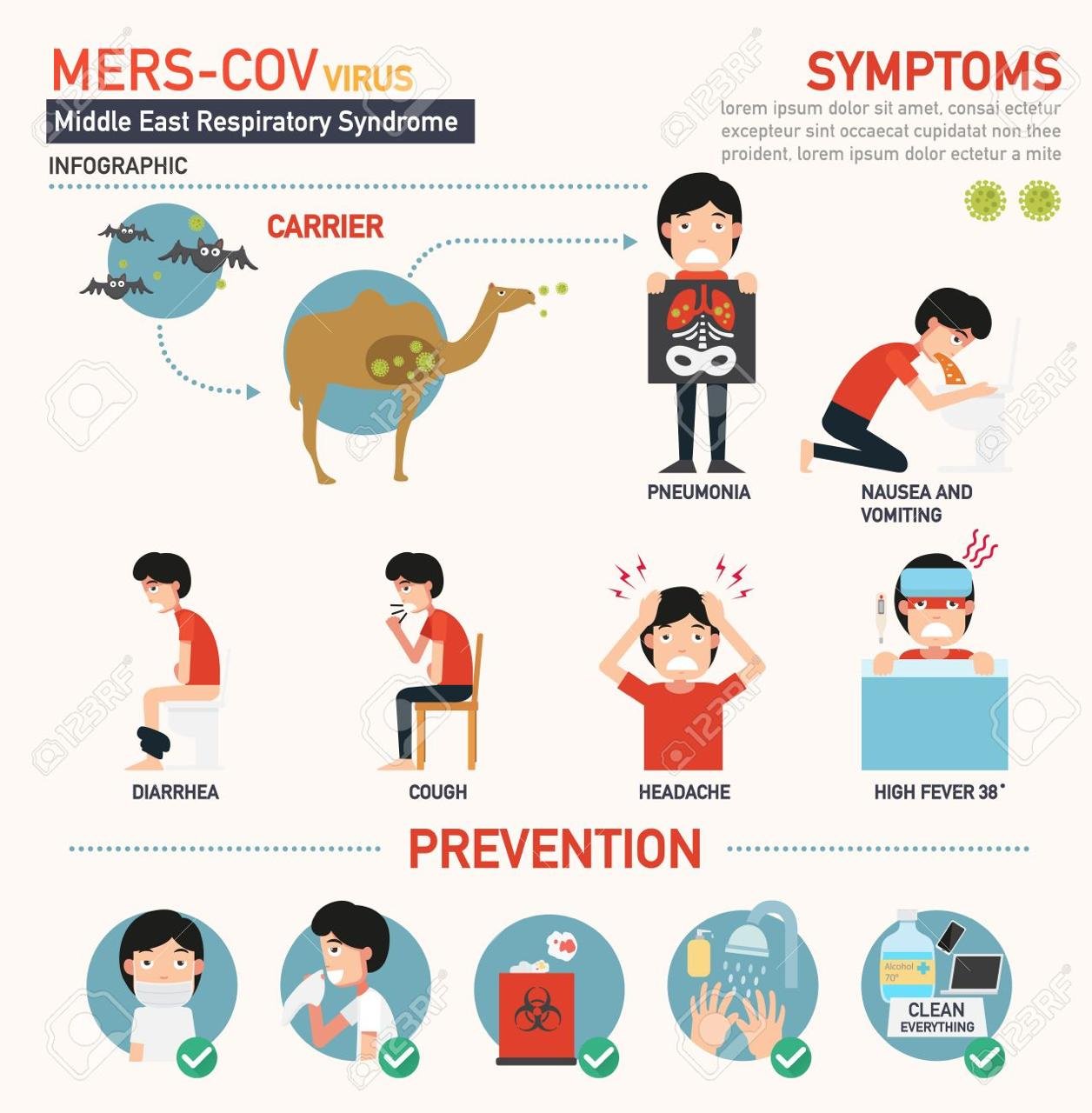
The largest known outbreak of MERS outside the Arabian Peninsula occurred in the Republic of Korea in 2015. The outbreak was associated with a traveler returning from the Arabian Peninsula. MERS-CoV has spread from ill people to others through close contact, such as caring for or living with an infected person. MERS can affect anyone. MERS patients have ranged in age from younger than 1 to 99 years old.
2019 Novel Human Corona Virus (2019 nCoV)
Please click here for further information.
The writer is a medical student.